Recent Structures
cMBD2:mCpG
We solved the structure of the chicken MBD2MBD bound to a methylated DNA sequence from the chicken ρ globin promoter (2KY8). The domain consists of a three-stranded β sheet which extends down the major groove to make base-specific contacts with DNA. A small C-terminal α-helix and N- and C- terminal loop regions pack against the opposite face of this β sheet. Two highly conserved arginine residues form bidentate hydrogen bonds with the symmetrically related guanosine bases of the mCpG dinucleotide. The aliphatic portion of the arginine side chains packs against the methyl groups of the preceding 5mC. A critical tyrosine residue points towards one 5mC such that the hydroxyl either interacts directly with the methyl group (forming a methyl hydrogen bond) or replaces a structured water molecule surrounding the methyl group.
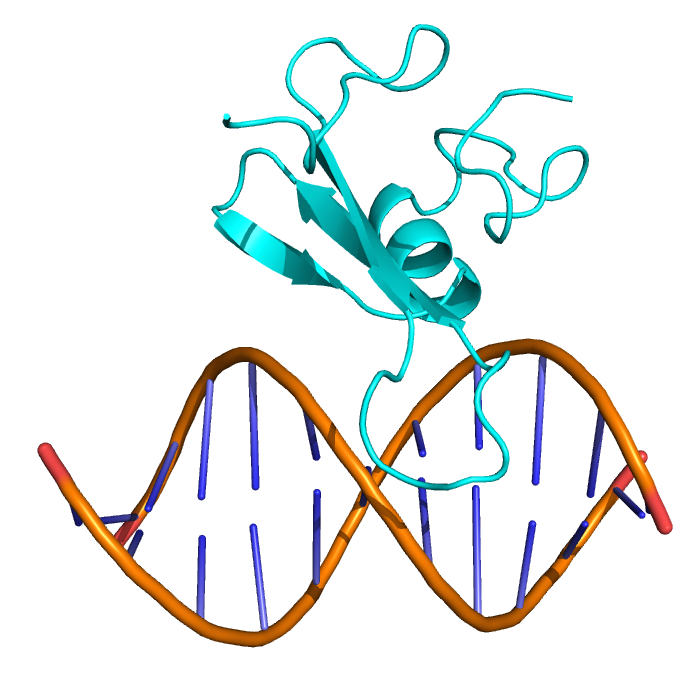
The MBD2MBD binds in the major groove of DNA. Two β-strands form a long finger-like projection that extends down the major groove and provides most of the contacts with DNA.
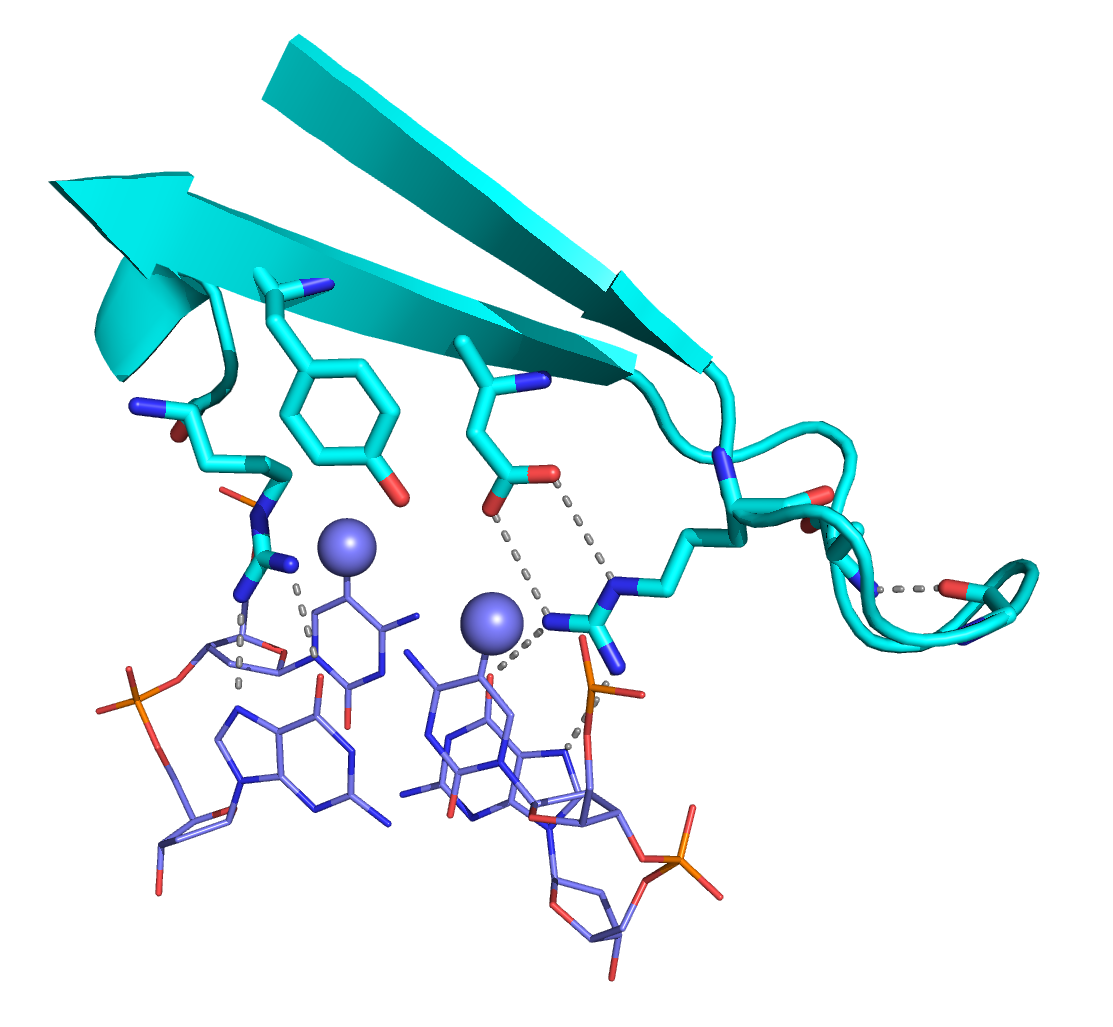
Two key arginine residues form bidentate hydrogen bonds with the symmetrically related guanosine bases of the CpG dinucleotide. A conserved aspartate residue stabilizes one arginine, while a tyrosine interacts with the methyl-cytosine.
p66α:MBD2 coiled-coil complex
Based on previous work we postulated and showed that the coiled-coil domains of MBD2 and p66α form a stable heterodimeric complex. These domains bind one another with high affinity (~20 nM) yet largely remained as monomers in isolation. We determined the solution structure of p66α:MBD2 (2L2L) which revealed a canonical anti-parallel coiled-coil complex with the typical “knobs into holes” hydrophobic interface stabilized by specific ionic interacions.
Based on this structure, we then showed that the p66α coiled-coil peptide could block the formation of the native NuRD complex and inhibit methylation-dependent gene silencing. This observation raises the possibility that a cell penetrating peptide or peptide mimetic could be developed for the treatment of cancer and β-hemoglobinopathies.
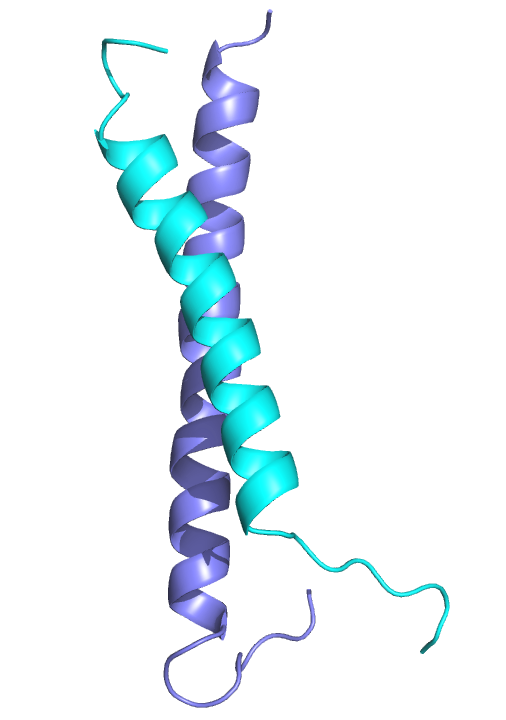
The MBD2 and p66α proteins from the NuRD complex form a canonical anti-parallel heterodimeric coiled-coil complex.
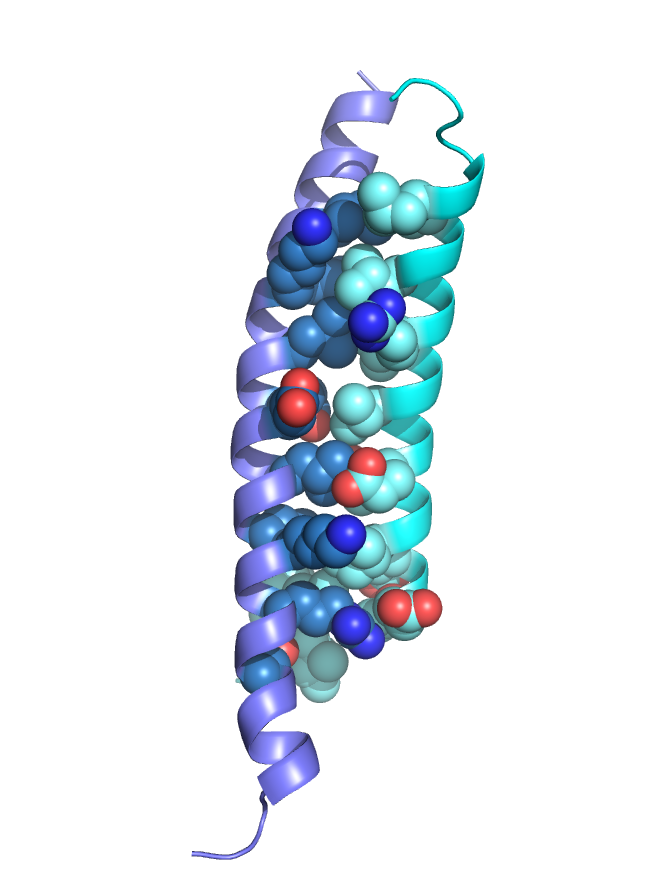
Hydrophobic “knobs” fit into “holes” as the two helices wrap around one another. Key charge interactions stabilize the complex and promote the anti-parallel arrangement.
MBD3:hmCpG
We recently solved the sructure of MBD3MBD bound to DNA with a central hydroxymethylated CpG dinucleotide (2MB7). The domain architecture is very similar to other MBDs (for comparison MBD3MBD is aligned to MBD2MBD on DNA). The critical sequence specific tyrosine has been changed to a phenylalinine in MBD3 leading to a lack of methylation selectivity. As might be expected from the relative lack of sequence specificity, we found that MBD3MBD did not localize to a single binding site on the DNA but distributed between (m)CpG specific and non-specific binding modes.
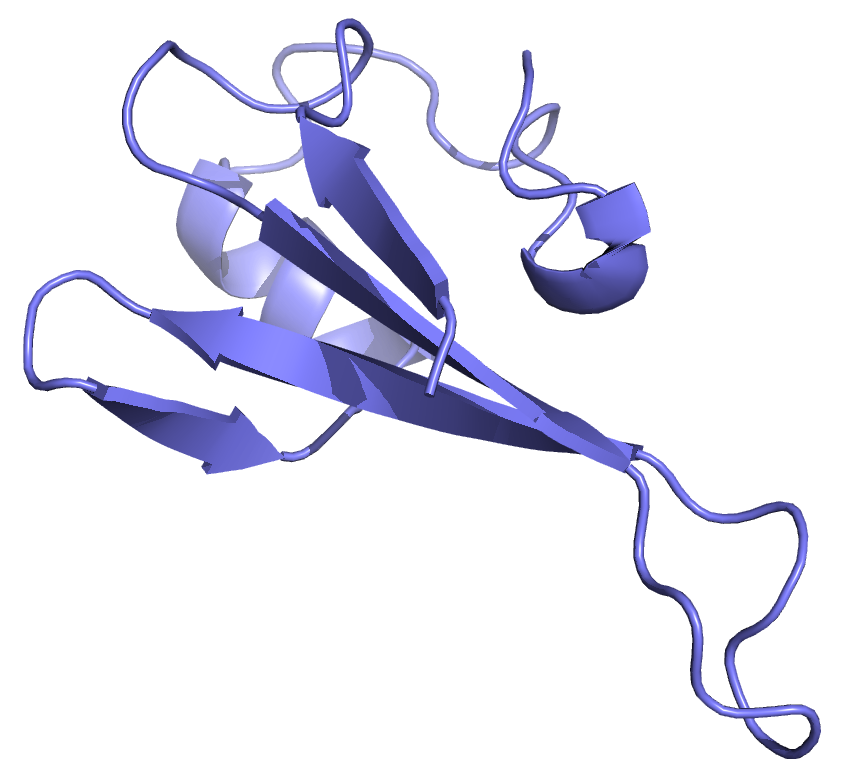
The MBD3MBD adopts a the typical MBD fold with the central two strands of a β-sheet forming a finger-like projection to extend down the major groove. NMR data, however, indicated that the domain exchanged rapidly between CpG specific and non-specific binding modes.
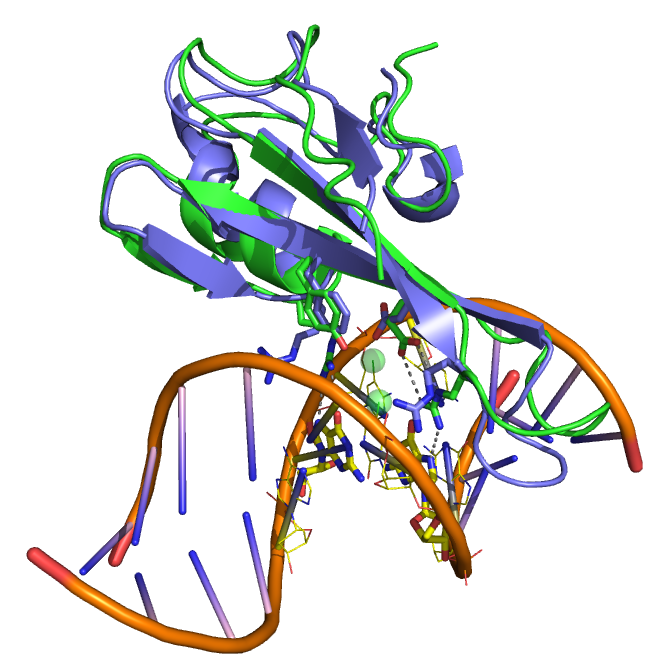
An overlay of MBD3MBD with MBD2MBD shows the structural similarity. A key difference between the two domains involves a tyrosine to phenylalanine change in MBD3 which reduces methylation specificity.
We used chemical shift and residual dipolar coupling analyses to characterize the distribution of MBD3MBD on DNA. We found that MBD3MBD preferentially localizes to methylated CpG sites as compared to unmethylated and hydroxymethylated CpGs. Chemical shift changes also indicated that MBD3 slightly favored CpG and hmCpG sites as compared to non-specific DNA interaction. Localization depends on the density of (m)CpG sites such that the fraction bound to a mCpG goes down as the length of the DNA is increased.
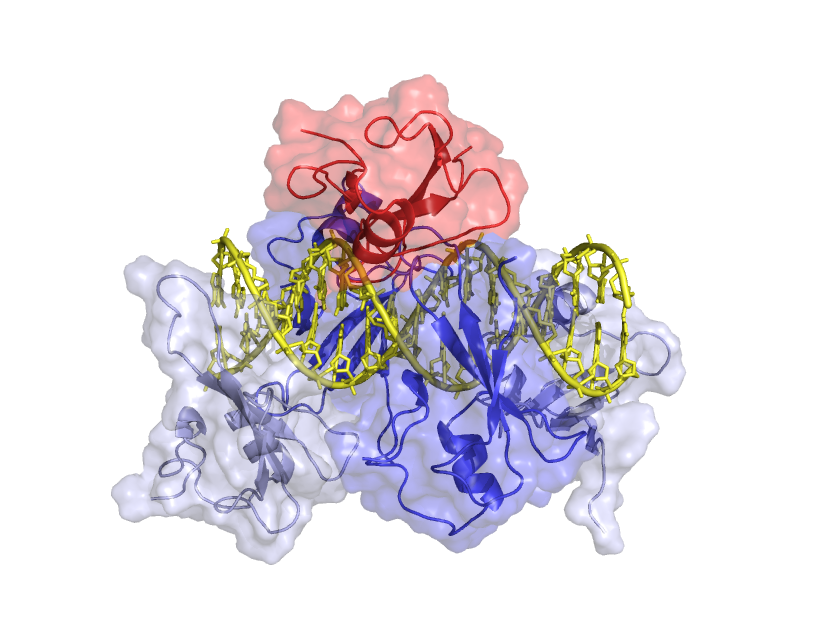
MBD3MBD exchanges between methylated and unmethylated sites along the DNA, spending more time on the methylated site.
These findings lead to a model in which MBD3MBD distributes unevenly along the DNA, favoring sequences that contain multiple methylated and unmethylated CpG sites. In contrast, MBD2MBD exclusively localizes to methylated CpGs when DNA is methylated and more strongly localizes to unmethylated CpGs as compared to MBD3MBD. This model of MBD2/3 distribution on DNA correlates with recent studies showing that both MBD2 and MBD3 localize to unmethylated CpG islands while MBD2 more exclusively localizes to methylated CpG islands.
MBD4:mCpG
More recently we determined the solution structure of MBD4MBD bound to methylated DNA (2MOE). Similar to MeCP2MBD. the MBD4MBD contains a small insertion that leads to a longer C-terminal α-helix and larger hydrophobic core. Unlike other MBDs, however, the key tyrosine residue in MBD4MBD does not interact with the mC but is rotated towards the phosphate backbone. In the figures to the right, the orientation of Tyr109 of MBD4 is compared to the equivalent Tyr123 of MeCP2 which does interact with the mC. This relatively subtle structural change in MBD4MBD leads to an overall reduced binding affinity and selectivity for methylated DNA.
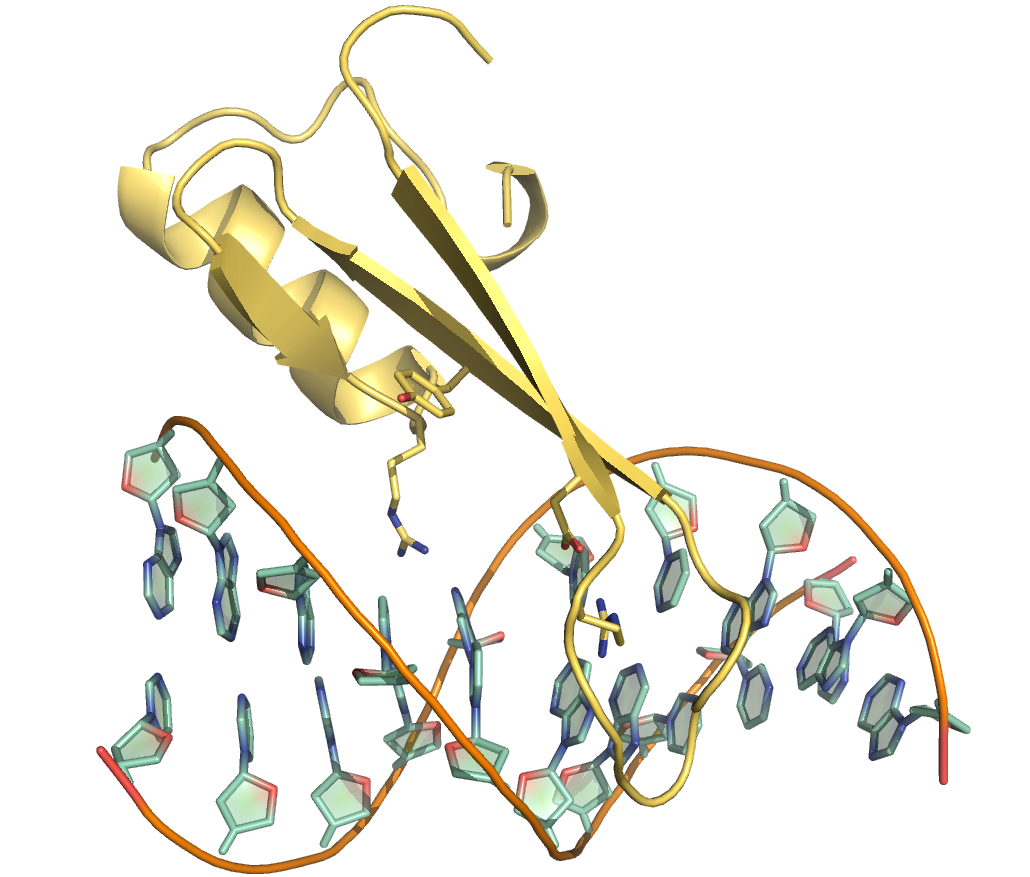
The MBD4MBD adopts a canonical MBD fold and binds to DNA with a similar orientation as MBD2MBD.
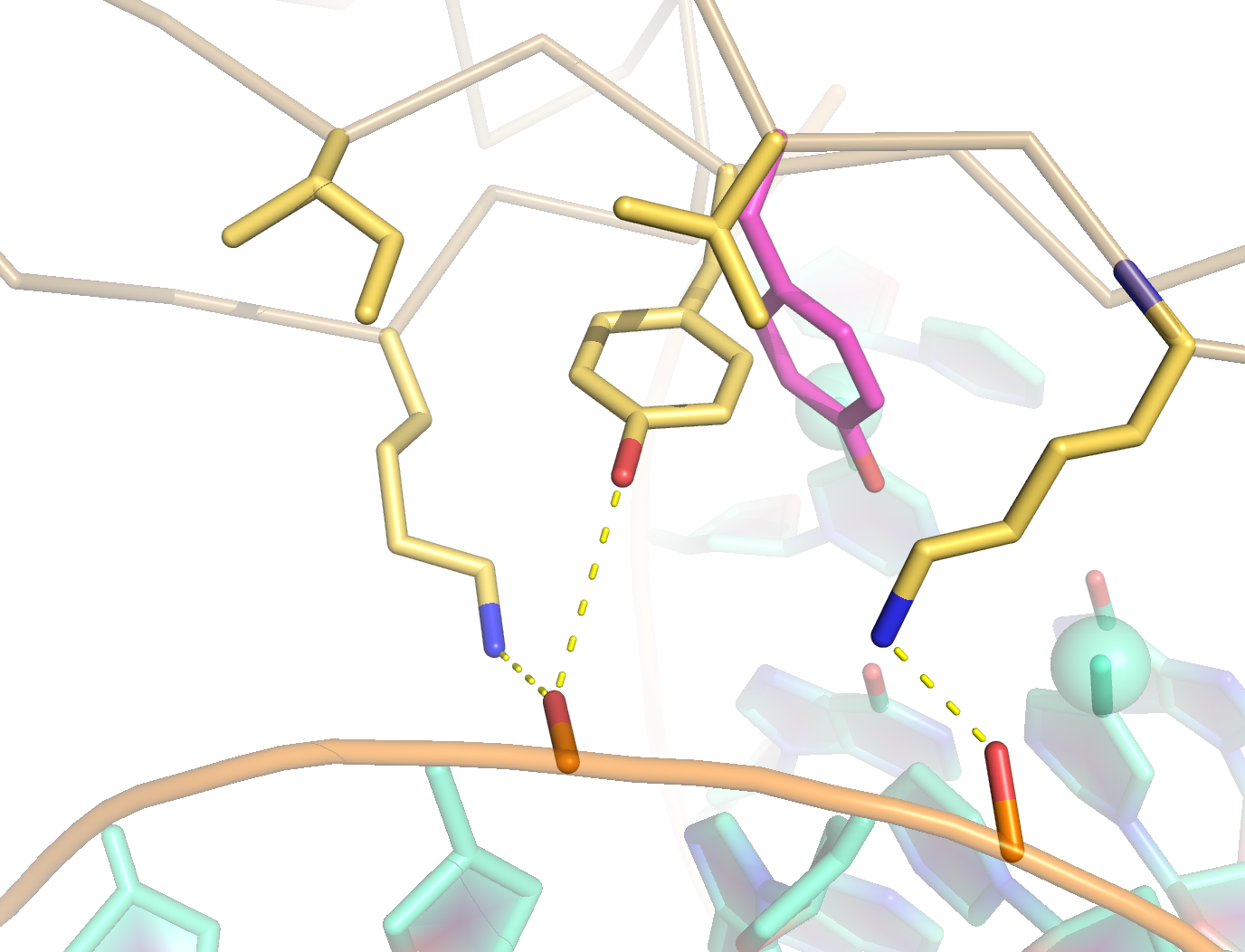
A key tyrosine residue adopts a unique conformation in MBD4MBD such that the sidechain points towards the phosphate backbone as opposed to interacting with the methyl-cytosine as in MeCP2MBD and other MBDs.
Unlike other members of the MBD family, MBD4 contains enzymatic activity within a C-terminal glycosylase domain (MBD4GD). The MBD4GD recognizes TpG·mCpG mismatches that arise from spontaneous deamination of mC. The N-terminal MBD4MBD is separated from the MBD4GD by approximately 300 amino acids which suggest that the MBD4MBD recruits the protein to methylated DNA and allows the MBDGD to identify and repair nearby mismatches. We have recently characterized inter- and intra-molecular exchange of MBD4MBD and found that this domain more efficiently exchanges between mCpG sites along the same DNA molecule as compared to separate DNA molecules. These observations are consistent with our model of MBD4MBD in which the primary function of this domain is to localize the protein to regions enriched for mCpG and then to rapidly exchange among binding sites.