Current Research
Dynamics and distribution of MBD proteins on DNA
- Why do MBD2 and MBD3 selectively localize to methylated and unmethylated CpG islands?
- How does methylation influence localization and exchange kinetics of the MBD proteins?
- Do the intrinsically disordered regions of MBD2 and MBD3 influence behavior on DNA?
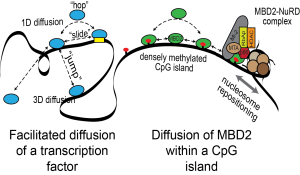
Transcription factors identify binding sites through a combination of free diffusion and 1D sliding, hopping, and jumping mechanisms. MBD2 binds to densely methylated CpG islands and recruits the NuRD chromatin remodeling complex. We hypothesize that rapid exchange between nearby CpG sites is critical to the function of MBD proteins.
Using the DNA tightrope assay developed by the Wang laboratory, we can visualize MBD2 diffusing along DNA (courtesy of Hai Pan, NC State University).
Ninad M. Walavalkar et al. Nucl. Acids Res. 2014;42:11218-11232 (modified) © The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
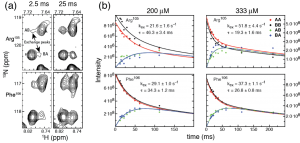
(a) Select resonances in the 2D 15N-HSQC of MBD4MBD show two distinct chemicals shifts when bound to a mixture of two unique mCpG sites. Cross peaks (AB and BA) between these resonances indicate chemical exchange between the two sites during the pulse sequence between t1 and t2.
(b) Inserting an additional delay between t1 and t2 during the HSQC pulse sequence while magnetization is along the Z axis allows for the cross peaks to build in intensity (Z-exchange spectroscopy). The rate of exchange between binding sites can be determined by fitting the intensities of AA, AB, BA, and AB cross peaks.
Recruitment and formation of NuRD
Building on this observation, we are now determining the molecular details of MBD2-NuRD complex formation using a combination of FRET/BRET, NMR, and x-ray crystallography.
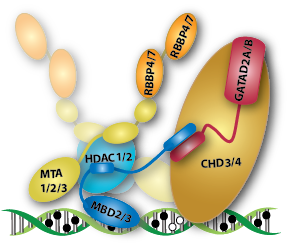
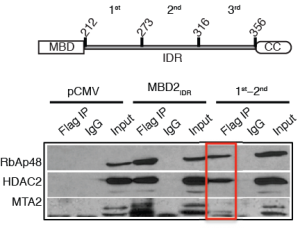
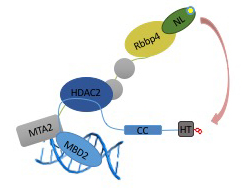
The NuRD complex can be divided into two functional and physical halves comprising the histone deacetylase core and the chromatin remodeling portions. The MBD2 IDR binds and recruits the MTA1/2, HDAC1/2, and RBBP4/7 components. The MBD2/3 coiled-coil domain binds GATAD2A/B which in turn binds the large CHD4 nucleosome remodeling protein.
Megha A. Desai et al. Nucl. Acids Res. 2015;43:3100-3113 (modified) © The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
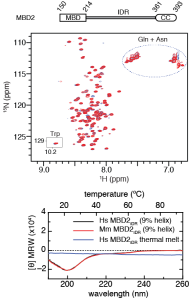
2D 15N-HSQC spectra and circular dichroism measurements of the MBD2 IDR are characteristic of a disordered peptide. Analytical ultracentrifugation shows that the region behaves largely as a monomer in isolation.
Megha A. Desai et al. Nucl. Acids Res. 2015;43:3100-3113 (modified) © The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
The first two-thirds of the MBD2 IDR can immunoprecipitate the histone deacetylase core complex of NuRD.
We are currently characterizing the structural and physical determinants of binding between MBD2 IDR and the histone deacetylase core complex. These studies involve a variety of biophysical techniques in vitro and in cells.
Resonance energy transfer between NanoLuc® (NL) bioluminescent domain and HaloTag® (HT) with an attached fluorophore.
Disrupting NuRD complex formation MBD2:GATAD2A coiled-coil complex
Studies have convincingly shown that disrupting MBD2 function restores expression of silenced genes while leading to only mild phenotypic changes. Therefore, MBD2 represents an exciting target for restoring expression of aberrantly methylated and silenced tumor suppressor genes for cancer as well as developmentally silenced genes such as γ-globin to treated inherited β-hemoglobinopathies (i.e. sickle cell anemia and β-thalassemia).
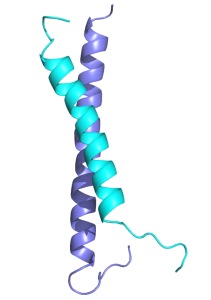
We previously solved the structure of a key coiled-coil interaction between the MBD2 and GATAD2A proteins in NuRD and showed that enforced expression of the GATAD2A coiled-coil peptide blocks methylation-dependent gene silencing by MBD2.ref
The MBD2 and GATAD2A proteins from the NuRD complex form a canonical anti-parallel heterodimeric coiled-coil complex.
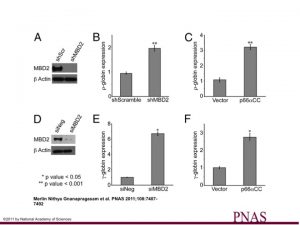
The GATAD2A coiled-coil peptide augments fetal/embryonic globin expression.
Building on that proof-of-principle experiment, we are developing cell-permeable peptides that can inhibit the MBD2-GATAD2A interaction in vitro and intracellularly, as well as the necessary assays to characterize its function in cells. This molecular probe will provide a powerful new tool to investigate the effects of this interaction on complex formation and gene silencing in the chromatin environment of cells, and how this MBD2-GATAD2A sub-complex interacts with other components of the intact NuRD.